HOW MANY COULD THERE POSSIBLY BE?
Identify and quantify threatening microbes
LifeCheck DNA qPCR is innovative genetic testing for rapidly determining problematic microbial populations. It enables the detection and accurate quantification of specific microbes without the need for growth media. Our qPCR technology can target specific microbial functional groups as well as individual species of interest. The reported results provide the total count of specific targeted microbes detected in the sample provided.
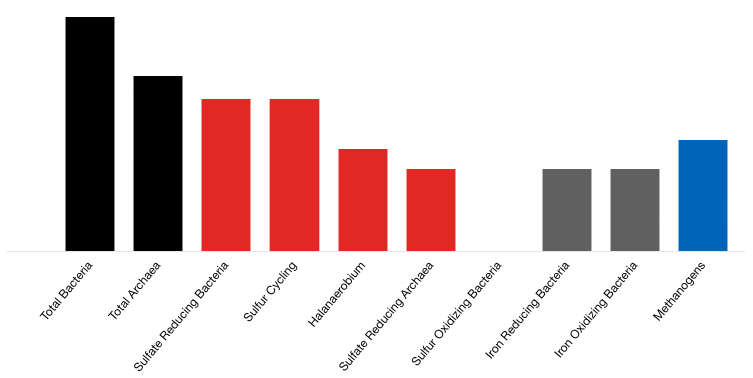
Sample Reported Results: Sour Producing Well
LifeCheck DNA qPCR Packages
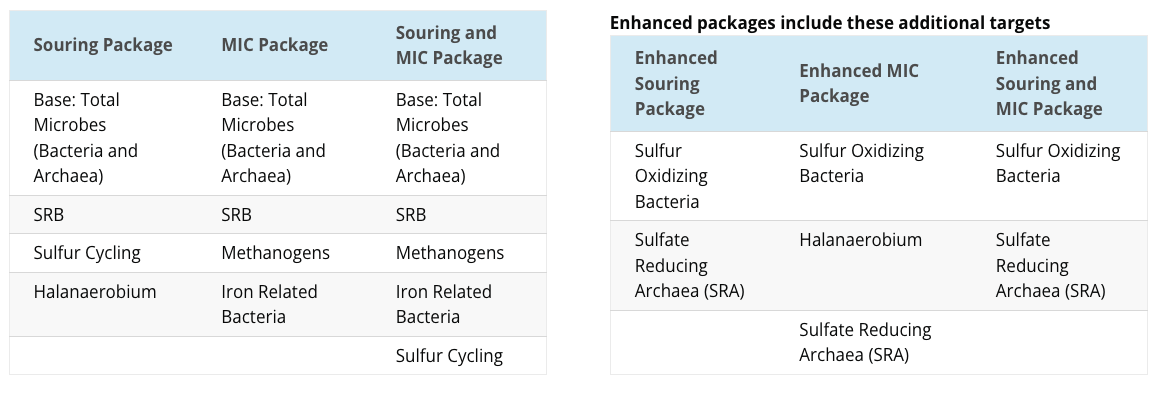
Understanding your LifeCheck DNA qPCR Package
qPCR Primer Set
Total archaea
A summary count of all the archaea in a sample.
Quality Controls
Internal DNA controls are used to ensure accurate and precise qPCR quantification.
Sulfate reducing bacteria
Sulfate reducing bacteria (SRB) gain energy for growth by reducing sulfate (SO42-) to sulfide (H2S). Sulfide production can cause souring of a system.
SRB can also cause MIC by directly removing electrons from steel surfaces, and indirectly by producing corrosive by-products such as sulfide which react with iron to produce iron sulfide (FeS) deposits.
Sulfate reducing archaea
Sulfate reducing archaea (SRA) gain energy for growth by reducing sulfate (SO42-) to sulfide (H2S). Sulfide production can cause souring of a system, and/or MIC.
Methanogens
These anaerobic Archaea produce methane during their metabolism.
Can lead to MIC by removing electrons directly from steel surfaces or indirectly through syntrophic interactions with other microbes.
Iron oxidizing bacteria
Iron oxidizing bacteria (IOB) gain energy for growth by oxidizing ferrous iron (Fe2+) to ferric iron (Fe3+), resulting in the formation of ferric iron oxides on the steel surface.
These oxides can be a protective physical barrier preventing any further corrosion, however they also result in the formation of oxygen depleted zones leading to electrochemical alterations of the steel surface, and localized, pitting corrosion.
Iron reducing bacteria
Iron reducing bacteria (IRB) gain energy for growth by reducing ferric iron (Fe3+) to ferrous iron (Fe2+).
Fe3+ reduction can remove protective oxide coatings, exposing the surface beneath to further corrosion.
Sulfur Cycling
A broad spectrum qPCR target evaluating the genetic capacity for microbial conversion of sulfur related compounds.
Supplementary to the total SRB qPCR primer set, the presence of “sulfur cycling” genes indicates a potential for microbial souring and/or MIC.
Sulfur Oxidizing Bacteria
A qPCR primer targeting the oxidation of sulfur-related compounds (sulfide, sulfur, thiosulfate) to sulfate.
Often considered a beneficial group of microbes as they function to remove sulfide (H2S) from a system, and are stimulated (used) in nitrate injection strategies for souring control. Recent research has indicated select SOB species can cause MIC by oxidizing sulfide to sulfuric acid (H2SO4).
Thiosulfate Reducing Bacteria
A sulfide producing microbe, increasingly common to oil and gas reservoirs stimulated by hydraulic fracturing methods.
Sulfide production occurs via the use of thiosulfate and not sulfate, making this microbe a non-traditional SRB, undetectable by culture media bottle methods.
Sulfide production poses both a souring and corrosion-based risk.
Are you killing or growing bugs?
Implementing a robust microbial monitoring and testing program that starts with your source water and follows through all the way to production, will let you know. At the end of the day the name of the game is control keeping the microbial load low enough and the environment, or conditions for growth, inhospitable enough that microbial communities can’t get a foothold and create a long term problem. Using a combination of ATP Testing and next level DNA testing provides the data and baseline KPI’s to ensure your control program is killing, not growing bugs.
Frequently Asked Questions
What is the difference between 16S Sequencing and qPCR?
- LifeCheck DNA 16S Sequencing characterizes and provides insight into the entire microbial community present, and in what relative proportion. Once the microbial community makeup is determined, a trained microbiologist can determine the types of functions (harmful activities) the microbial community may be capable of.
- LifeCheck DNA qPCR is innovative genetic testing that allows the user to target and accurately quantify specific subgroups or types of microbes without the need for growth media.
