TIME TO LINE UP THE SUSPECTS AND FIND OUT WHO THE REAL CULPRITS ARE
Evaluate problematic microbes present with 16S Sequencing
Features and Benefits

ACCURATE
No growth biases, no false negatives or false positives.

INFORMATIVE
Data sent in an easy to understand report. Make informed mitigation decisions.

Customizable Essays Data
LAB QUALITY ASSAYS
OSP employs multiple QC and QA controls throughout the DNA extraction and testing processes to ensure the highest level of quality throughout the entire process.

Lifecheck DNA 16S Sequencing
ROBUST
It’s no secret, oil and gas related samples are some of the most difficult to extract DNA from. With its fully equipped molecular lab, OSP is able to extract quality DNA from almost any sample, and clean the most difficult of samples from contaminants.

Easy Analysis
EASY
OSP provides end users with easy to use sampling kits. These kits contain a chemical preservative that arrests changes to the microbial community, ensuring the most representative sample is studied.
Did you know?
Most microbes are not easily grown in culture media, which have been shown to grow less than 1% of known microbes in the environment. This means that important decisions are being made about microbial control and corrosion mitigation programs without 99% of the required information. You can’t mitigate what you can’t measure!
Because LifeCheck DNA is not a growth-based test, accurate results can be ready in days instead of weeks as with traditional culture methods. It works well together with the LifeCheck ATP test, which determines the total active (living) microbes in a sample. The LifeCheck DNA test will then tell you what proportion are problematic microbes (e.g. SRB, methanogens, iron related microbes).
Use DNA technologies for in depth insights and problem solving.
Here are a few things to think about when setting up a testing program:
- Is my biocide program working?
- Have I seen growth in my system that needs to be investigated?
- Why am I having treatment issues?
- What is my baseline microbial load?
Programs require consistency in testing frequency, location and method. They often necessitate trending so changes can be evaluated. Additionally, don’t forget that when one test method doesn’t satisfy your objectives, there are more options.
Frequently Asked Questions
LifeCheck DNA technology is a modern microbial identification and evaluation tool for determining problematic microbial populations in almost any sample type utilizing either 16S rDNA sequencing or qPCR. This innovative genetic testing works by measuring the amount of specific DNA present in the sample from microbes of interest. LifeCheck DNA can provide a full microbial community profile, or quantify targeted microbes.

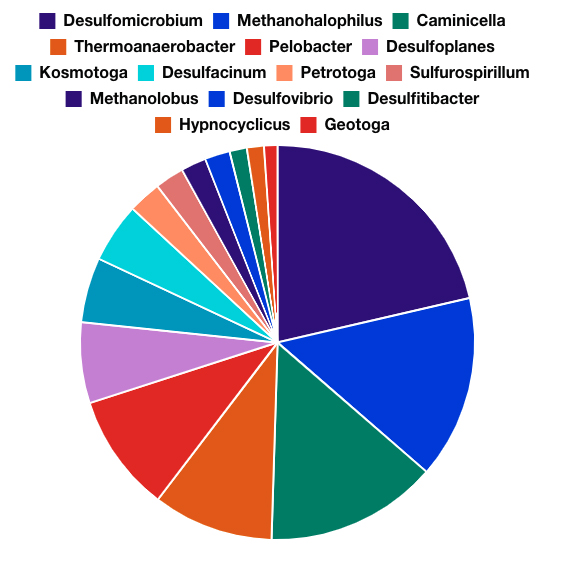
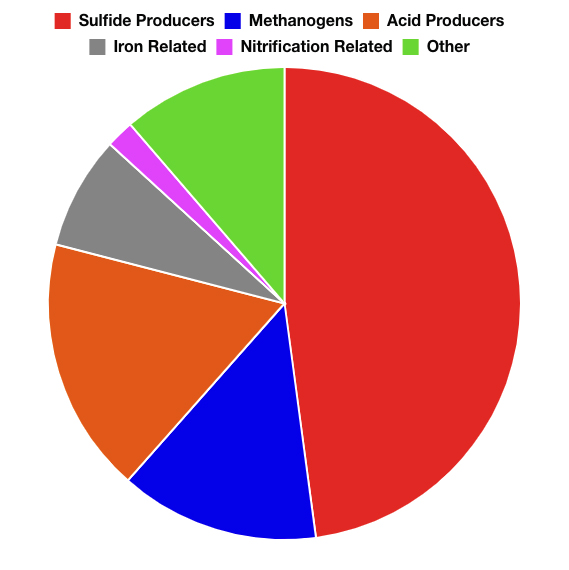
- LifeCheck DNA 16S Sequencing characterizes and provides insight into the entire microbial community present, and in what relative proportion. Once the microbial community makeup is determined, a trained microbiologist can determine the types of functions (harmful activities) the microbial community may be capable of.
- LifeCheck DNA qPCR is innovative genetic testing that allows the user to target and accurately quantify specific subgroups or types of microbes without the need for growth media.